Rather than creating de novo structures out of thin air (like a greek God), protein engineering usually consists of slightly mutating natural proteins to give them more desired properties.
But what is meant by mutating and what is the desired result?
We might be getting ahead of ourselves, so let's do a refresher on how proteins work. To understand all of this you first have to know the central dogma of biology. DNA (the longterm code that determines our genetics) is transcribed into RNA (short lived code), which is translated into proteins. Proteins help build up the structure and carry out the biological functions of organisms.
A protein is made up of one or more polypeptide chain subunits. Polypeptide chains are made from linking a choice of twenty amino acids (smallish molecules) end-to-end. These subunits fold together under normal (normal = physiological, i.e inside the body) pH and temperature conditions into a specialized structure. This specialized structure performs a certain function in a cell or organism.
Always remember, in Biology, the form of an object is associated with the function of the object. Structure is everything.
When building a protein there are 20 amino acids to choose from:
1. Glycine, (Gly, G)
2. Alanine (Ala, A)
3. Valine (Val, V)
4. Leucine (Leu, L)
5. Isoleucine (Ile, I)
6. Tryptophan (Trp, W)
7. Phenylalanine (Phe, F)
8. Tyrosine (Tyr, Y)
9. Threonine (Thr, T)
10. Serine (Ser, S)
11. Cysteine (Cys, C)
12. Methionine (Met, M)
13. Glutamatic Acid/Glutamate (Glu, E)
14. Glutamine (Gln, Q)
15. Arginine (Arg, R)
16. Aspartic Acid/Aspartate (Asp, D)
17. Asparagine (Asn, N)
18. Lysine (Lys, K)
19. Histidine (His, H)
20. Proline (Pro, P)
 |
| The general form for an amino acid. |
An amino acid has four basic parts surrounding a central alpha carbon: an amine group, a carboxyl group, a Hydrogen, and a wild card R group. The carboxylic group of one amino acid links with a following amino acids' amide group to form a peptide bond. A series of these peptide bonds forms a peptide chain. This is a protein's primary structure. If our protein was a piece of clothing, we might say that its primary structure is a single strand of thread that makes up the clothing.
 |
| An Alpha Helix |
The chain will then fold into what is called secondary structure. The most common secondary structure types are the alpha helix and the beta sheet. The alpha helix looks like a right handed stair case that twists upwards by 3.5 residues per turn. The R groups of each residue (amino acid) stick out the sides of this long winding twist. The beta sheets look like wavy sheets stacked on top of each other. For our clothing metaphor, the secondary structure might be small knots and loops in the thread.
 |
| A beta sheet |
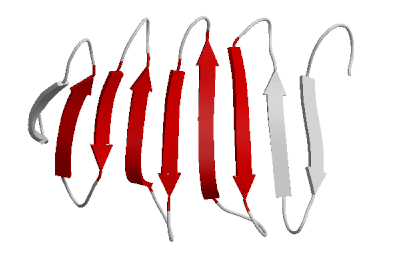 |
| Another representation of a beta sheet. |
The tertiary structure is the overall 3D globular structure of a protein. This is the point where proteins really start looking like, well, proteins. There is a 3D structure. For our clothing metaphor, you can imagine the tertiary structure is the formation of a pant leg. Some proteins only go up to this tertiary structure.
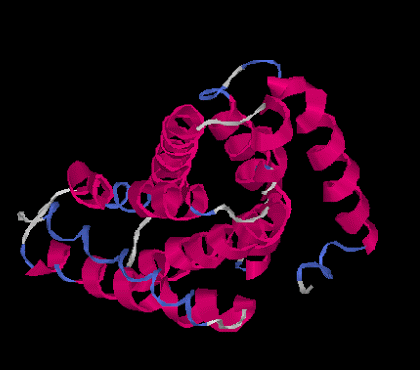 |
| Tertiary Structure |
Sometimes, but not always, tertiary structure pieces made of different peptide chains come together to form quaternary structure. You can think of this as pant legs and other pieces coming together to make a pair of functional jeans. The pieces are from a different strand of thread (a different peptide chain), but the parts work together.
 |
| Hemoglobin is protein with Quaternary structure. |
Sources:
https://upload.wikimedia.org/wikipedia/commons/7/74/Alpha-amino-acid-general-2D.png\
https://upload.wikimedia.org/wikipedia/commons/7/75/Alpha_helix.png
https://upload.wikimedia.org/wikipedia/commons/thumb/c/c9/Alpha_sheet_bonding_schematic-color.svg/1004px-Alpha_sheet_bonding_schematic-color.svg.png
https://upload.wikimedia.org/wikipedia/commons/b/b8/Beta-meander1.png
https://upload.wikimedia.org/wikipedia/commons/2/2d/I-Tasser_Predicted_Tertiary_Structure_1.png
https://upload.wikimedia.org/wikipedia/commons/2/26/225_Peptide_Bond-01.jpg
https://upload.wikimedia.org/wikipedia/commons/7/75/Alpha_helix.png
https://upload.wikimedia.org/wikipedia/commons/thumb/c/c9/Alpha_sheet_bonding_schematic-color.svg/1004px-Alpha_sheet_bonding_schematic-color.svg.png
https://upload.wikimedia.org/wikipedia/commons/b/b8/Beta-meander1.png
https://upload.wikimedia.org/wikipedia/commons/2/2d/I-Tasser_Predicted_Tertiary_Structure_1.png
https://upload.wikimedia.org/wikipedia/commons/2/26/225_Peptide_Bond-01.jpg
